Abstract
The association of positive sepsis PCRs tests with patient mortality, length of stay, and antibiotic management in a medical intensive care unit
Ximena Solis MD, Ben Batson DO, Cristina Morataya MD, MPH, Hannah Fairley MD, Wadih Chakkour MD, Kenneth Nugent MD, Ebtesam Islam MD, PhD
Corresponding author: Ebtesam Islam
Contact Information: Ebtesam.Islam@ttuhsc.edu
DOI: 10.12746/swrccc.v11i49.1231
ABSTRACT
Background: Polymerase chain reaction (PCR) testing amplifies a specific DNA segment through heat cycling to identify pathogens for the diagnosis of infection. This testing allows for rapid detection of pathogens, before conventional blood culture results become available. The high mortality associated with severe sepsis and septic shock stresses the importance of early diagnosis and initiation of early antibiotic therapy. This study analyzed the association of the sepsis PCR results and antibiotic management, length of stay, and mortality outcomes in a medical intensive care unit (MICU).
Methods: This study is a retrospective, cross-sectional study on patients diagnosed with severe sepsis and septic shock based on ICD-10 codes, aged 18 years and older, admitted to the MICU at University Medical Center, Lubbock, Texas, between December 2016 to December 2020.
Results: Clinical information from 268 patients with the diagnosis of sepsis or septic shock was collected and analyzed. The mean age was 60.9 ± 15.6 years with a predominance of men (144, 53.7%) and Caucasian race (193,73.4%). A total of 101 patients (37.6%) had positive PCR results; overall, PCR test results had a diagnostic sensitivity of 91.2 %. The concordance between positive blood culture and positive PCR was 93.3 % (p < .0001). There was a significant correlation between PCR positivity and increased serum lactate levels (p = 0.03), changes in antibiotics (P < 0.00010), and increased mortality rate (p = 0.04). There was no significant correlation between PCR positivity and length of stay (p = 0.84).
Conclusion: In this study, PCR testing was an accurate tool for early identification of bacterial pathogens. A positive PCR was associated with higher serum lactate levels, higher mortality rates, and an increased frequency of antibiotic changes but was not associated with shorter ICU length of stay.
Keywords: Sepsis, septic shock, PCR testing, outcomes
INTRODUCTION
Sepsis continues to represent an important disorder in intensive care medicine given its healthcare burden and associated high mortality rates.1 Prompt and accurate diagnosis of sepsis is crucial to improve survival. Therefore, several guidelines have been updated in recent years to better define this term. The Third International Consensus Definition for Sepsis and Septic Shock or Sepsis 3 revised and updated sepsis and septic shock definitions in 2016.2 Severe sepsis is defined as sepsis with organ dysfunction. Organ dysfunction can be measured using the Sequential Organ Failure Assessment or SOFA score and is defined by an acute change of 2 points or more. Severe sepsis is associated with a hospital mortality greater than 10%. Septic shock is the progression of severe sepsis in which organ dysfunction remains despite fluid resuscitation. It is defined as having a vasopressor requirement to keep mean arterial pressure higher than 65 mmHg and having a serum lactic acid level greater than 2 mmol/L in the absence of hypovolemia. Septic shock is associated with hospital mortality greater than 40%.2
Currently, the gold standard for diagnosis of infection is a blood culture.3 However, not all pathogens are detected by blood cultures, and their value as the gold standard has been questioned with the development of new molecular techniques, such as polymerase chain reaction (PCR) testing, which can identify non-viable bacteria and non-culturable bacteria not identified on routine laboratory cultures.4 These tests amplify specific DNA segments through heat cycling to identify pathogens for the diagnosis of infection. Detection rates of both PCR and culture have been reported to be significantly higher than either method alone.5,6 In studies comparing PCR-based diagnostics with conventional blood cultures, more pathogens were detected by PCR techniques.3,5,6 In addition, PCR allows for rapid detection of slow-growing pathogens and can provide results in 6 hours vs. 48 hours with blood cultures.3
The high mortality associated with severe sepsis and septic shock stresses the importance of early diagnosis and initiation of early goal directed therapy. Therefore, we analyzed the diagnostic utility of the sepsis PCR and its association with antibiotic management, length of stay, and mortality outcomes in a medical intensive care unit (MICU).
METHODS
This study is a retrospective, cross-sectional study of patients aged 18 years and older, admitted to the MICU at University Medical Center in Lubbock, Texas, between December 2016 to December 2020. The inclusion criteria required a diagnosis of severe sepsis or septic shock based on ICD-10 code R65.20 for severe sepsis without septic shock and the ICD- 10 code R65.21 for severe sepsis with septic shock. Patients younger than 18 years and older than 89 years and patients with other sources of shock were excluded. This study was approved by the Institutional Review Board (L19-101) at Texas Tech University Health Sciences Center in Lubbock, Texas.
Demographic covariates of gender, race, and age were collected. Gender and race were defined as categorical variables with values for male and female, and values for Caucasian, African American, Hispanic, Asian, and other, respectively. Age was measured as a continuous variable in years. An additional severe sepsis-related continuous covariate was the serum lactate levels in mmol/L. The primary outcome was to determine the percent positive sepsis PCR tests reported with positive blood cultures. Secondary outcomes included determining the association between positive sepsis PCRs tests and mortality and MICU length of stay. For primary outcome variables, sepsis PCR status and blood culture status were analyzed as categorical variables with values of yes/no, in which a “no” result indicates that no PCR or no blood culture was done. The secondary outcomes of mortality and change in antibiotics were evaluated as categorical variables (yes/no), while length of stay in the MICU and time to antibiotic change were measured as numerical variables in days and hours, respectively.
GenMark® molecular rapid diagnostic technology (GenMark Diagnostics, Inc, Carlsbad, CA) was used for identifying bloodstream infections in this study. This system detects and identifies Gram-positive, Gram-negative, and fungal organisms using individual panels for the identification of each subgroup of microbial pathogens; this technology can also identify some antibiotic resistance genes.7 Gram staining is performed after a blood culture bottle is flagged positive by the production of CO2 during incubation. Then, the appropriate panel is selected based on the Gram stain results of the detected organisms. The reported timeframe for organism identification using this technology is 90 minutes after the blood culture flags positive. In a meta-analysis of 31 studies and 5,920 patients, the use of molecular rapid diagnostic technology was significantly associated with a decrease of mortality risk in the presence of an antibiotic stewardship program (odds ratio: 0.64; 95% CI: 0.51–0.79).8 In our study, only patients with a diagnosis of sepsis or septic shock with a positive blood culture had a subsequent PCR test done.
STATISTICAL ANALYSIS
All analyses were conducted in SAS 9.4 version (SAS Institute Inc, 2016) for Windows. The distributions of covariates in the dataset were reported as means (standard deviations) for continuous covariates and frequencies (percentages) for categorical covariates. Differences in covariates between patients with a positive PCR result and with a no PCR testing, were compared with independent t-tests or Wilcoxon rank sum tests for continuous variable analysis and Chi-square or Fisher’s exact test for categorical variable analysis. P values < 0.05 were considered statistically significant. Pearson’s correlation analysis was used to assess any relationship between PCR status and blood cultures and between PCR status with the secondary outcomes of LOS, lactate level, mortality rate, and change of antibiotics. In the Results section, a negative PCR means that no PCR test was done.
RESULTS
A total of 346 patients aged 18 years or older with a diagnosis of severe sepsis or septic shock based on ICD-10 codes were eligible for analysis. After removing patients with incomplete information and duplicates, the total analytic sample included 268 patients (Figure 1).
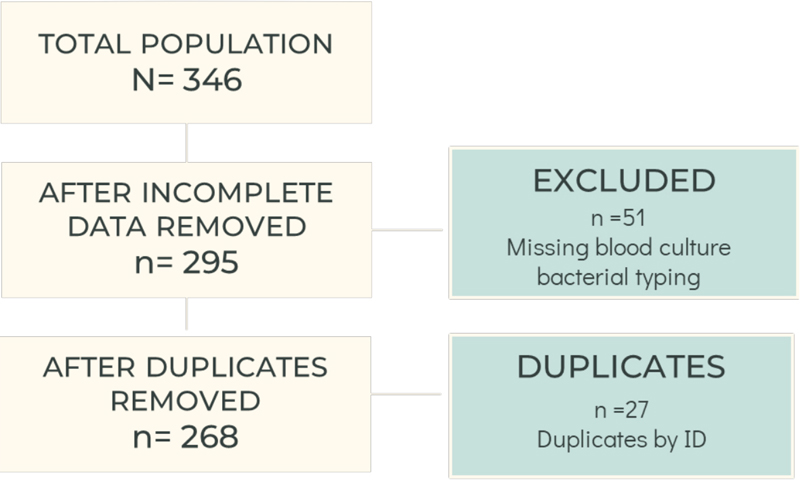
Figure 1. Analytic sample.
The mean age was 60.9 ±15.6 years with a predominance of male gender (144, 53.7%) and Caucasian race (196, 73.4%), followed by African American race (32, 12%), and Hispanic race (31, 11.6%) (Table 1). According to sepsis subtype, 38 patients (14.2%) had severe sepsis without septic shock, and 230 patients (85.8%) had severe sepsis with septic shock. Among the 268 patients with diagnoses based on ICD-10 codes, 101 patients (37.7%) had positive PCR results, and 114 patients (42.5%) had positive blood culture results (Table 2).
Table 1. Characteristics of Study Patients
| Variable |
Analytic Sample
n = 268 |
| Age, years, mean (SD) |
60.9 (15.6) |
| Race, # (%) |
|
| Caucasian |
196 (73.4) |
| African American |
32 (12.0) |
| Hispanic |
31 (11.6) |
| Other |
8 (3.0) |
| Gender, male, # (%) |
144 (53.7) |
| Severe sepsis, # (%) |
|
| No septic shock |
38 (14.1) |
| Septic shock |
230 (85.8) |
| PCR, yes, # (%) |
101 (37.6) |
| Time to antibiotic, hours, mean (SD) |
4.3 (9.9) |
| Change in antibiotic yes, # (%) |
126 (47.0) |
| Serum lactate level, mmol/L, mean (SD) |
3.3 (3.0) |
| LOS*, days, mean (SD) |
6.3 (6.0) |
| Mortality, # (%) |
89 (33.2) |
| Blood culture, yes, # (%) |
114 (42.5) |
| Infection sites, total (%) |
|
| Blood |
12 (4.5) |
| Pulmonary |
55 (20.6) |
| Urinary |
38 (14.2) |
| Abdominal |
16 (6.0) |
| Meningitis |
3 (1.1) |
| Joint |
4 (1.50) |
| Other |
28 (10.5) |
| Multiple |
111 (41.6) |
Table 2. Characteristics of Patients by PCR Status
| Variable |
Negative PCR *
n = 168 (63%) |
Positive PCR
n = 100 (37%) |
P value |
| Age, years, mean (SD) |
62.2 (16.4) |
58.9 (13.7) |
0.06 |
| Race, # (%) |
|
|
0.25 |
| Caucasian |
127 (76.1) |
69 (69.0) |
|
| African American |
17 (10.2) |
15 (15.0) |
|
| Hispanic |
16 (9.6) |
15 (15.0) |
|
| Other |
7 (4.2) |
1 (1.0) |
|
| Gender, male, # (%) |
92 (54.8) |
52 (52.0) |
0.66 |
| Severe sepsis, # (%) |
|
|
0.67 |
| No septic shock |
25 (14.9) |
13 (13.0) |
|
| Septic shock |
143 (85.1) |
87 (87.0) |
|
| Code sepsis, yes, # (%) |
47 (28.0) |
37 (37.0) |
0.12 |
| Time to antibiotic, hours mean (SD) |
4.4 (11.2) |
3.0 (4.3) |
0.32 |
| Change in antibiotic, yes # (%) |
62 (36.9) |
64 (64.0) |
<.0001 |
| Serum lactate level mmol/L, mean (SD) |
3.3 (3.3) |
3.5 (2.5) |
0.03 |
| LOS**, days, mean (SD) |
6.9 (7.2) |
7.0 (7.0) |
0.84 |
| Mortality, # (%) |
48 (28.6) |
41 (41.0) |
0.04 |
| Infection sites, total (%) |
167 (62.6) |
100 (37.5) |
<.0001 |
| Blood |
3 (1.8) |
9 (9.0) |
| Pulmonary |
50 (29.9) |
5 (5.0) |
| Urinary |
34 (20.4) |
4 (4.0) |
| Abdominal |
14 (8.4) |
2 (2.0) |
| Meningitis |
3 (1.8) |
0 |
| Joint |
3 (1.8) |
1 (1.0) |
| Other |
25 (15.0) |
3 (3.0) |
| Multiple |
35 (2) |
76 (76.0) |
Compared to no PCR results, positive PCR test results were significantly associated with more frequent antibiotic changes (64 changes [64.0%] vs. 62 changes [36.9%], P <.0001) and higher lactate levels (3.51 mmol/L vs. 3.28 mmol/L, P = 0.03). Compared to patients with no PCR testing, patients with positive PCR tests had a significantly higher mortality (41 deaths [41.0%] vs. 48 deaths [28.6%], p = 0.04). However, there was no significant difference between PCR status and length of stay (p = 0.84) or start time for antibiotic administration (p = 0.32) (Table 2).
Compared to having a negative PCR status, the most common site of infection in patients with positive PCR was bloodstream (9, 9%) and combined infection sites (76, 76%); patients with no PCR test results had more pulmonary infections (50, 29.9%), followed by combined infection sites (35, 21.0%), and urinary infections (34, 20.4%). The most frequent combined sites of infection in patients with a positive PCR test result were blood and urinary (29%) followed by blood and pulmonary (9%) (Figure 2).
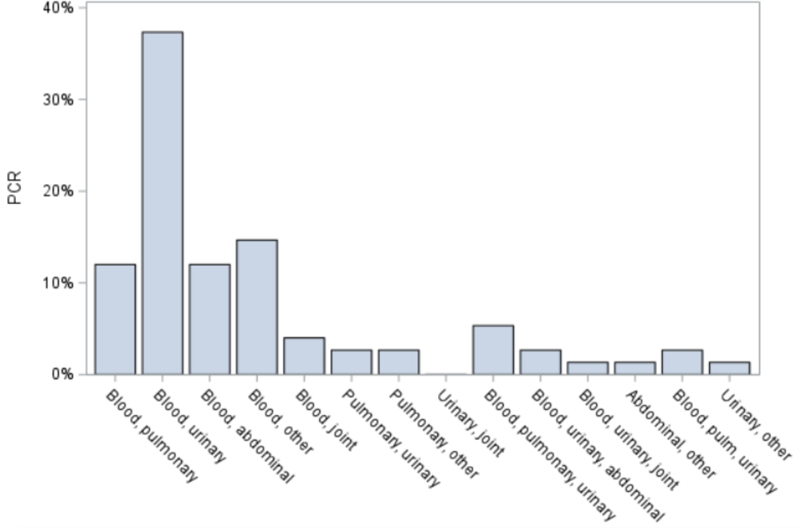
Figure 2. Percentage of combined infection sites in patients with a positive PCR test.
Patients with septic shock (230) had a 36.5% mortality rate; patients with severe sepsis (38) had a 13.2% mortality rate (P = 0.005) (Table 3). The diagnosis of septic shock was associated with a shorter time to the start of antibiotics of 4.2 ± 9.4 hours in comparison to 5.3 ± 12.3 hours for the diagnosis of severe sepsis (P = 0.02).
Table 3. Characteristics of Patients Classified by Shock Type
| Variable |
Severe Sepsis
n = 38 (14%) |
Septic Shock
n = 230 (86%) |
P value |
| Age, years, mean (SD) |
58.4 (20.5) |
61.3 (14.7) |
0.42 |
| Race, # (%) |
|
|
0.94 |
| Caucasian |
29 (76.3) |
167 (72.9) |
|
| African American |
5 (13.2) |
27 (11.8) |
|
| Hispanic |
3 (7.9) |
28 (12.2) |
|
| Other |
1 (2.6) |
7 (3.1) |
|
| Gender, male, # (%) |
14 (36.8) |
130 (56.5) |
0.02 |
| PCR, yes, # (%) |
13 (34.2) |
87 (37.8) |
0.67 |
| Time to antibiotic, hours mean (SD) |
5.3 (12.3) |
4.2 (9.4) |
0.02 |
| Change in antibiotic, yes # (%) |
19 (50.0) |
107 (46.5) |
0.78 |
| Serum lactate level mmol/L mean (SD) |
3.2 (2.3) |
3.3 (3.1) |
0.76 |
| LOS* (days) |
4.7 (4.4) |
4.2 (9.4) |
0.84 |
| Mortality, # (%) |
5 (13.2) |
84 (36.5) |
0.005 |
| Infection sites, total # (%) |
37 (13.9) |
230 (86.1) |
0.53
|
| Blood |
0 (0.00) |
12 (5.2) |
| Pulmonary |
10 (27.0) |
45 (19.6) |
| Urinary |
3 (8.1) |
35 (15.2) |
| Abdominal |
3 (8.1) |
13 (5.7) |
| Meningitis |
0 (0.0) |
3 (1.3) |
| Joint |
0 (0.0) |
4 (1.74) |
| Other |
6 (16.2) |
22 (9.6) |
| Multiple |
15 (40.5) |
96 (41.7) |
Overall, PCR status had a diagnostic sensitivity of 91.2%. Specificity could not be calculated with this data set since blood cultures with no growth would not have PCR tests run. The Pearson correlation between positive blood culture and positive PCR was 92.6% (P <.0001). Of 114 patients with positive PCRs, 85 patients (74.6%) had matching organisms in their blood cultures, and 29 patients (25.4%) had discordant results. Of the 29 patients with discordant results, 19 patients had different bacteria identified by PCR tests compared to blood cultures, and 10 had bacteria identified by blood culture but not by PCR, possibly explained by bacteria not on the PCR panel (Table 4 and Tables 5A and 5B).
Table 4.
|
Blood Cultures |
| PCR |
|
Positive |
Negative* |
Total |
| Positive |
104 |
0 |
104 |
| Negative |
10 |
154 |
164 |
| Total |
114 |
154 |
268 |
Table 5A.
|
Sepsis Blood Cultures |
| PCR |
|
Positive |
Negative* |
Total |
| Positive |
14 |
0 |
14 |
| Negative |
1 |
23 |
24 |
| Total |
15 |
23 |
38 |
Table 5B.
|
Shock Blood Cultures |
| PCR |
|
Positive |
Negative* |
Total |
| Positive |
90 |
0 |
90 |
| Negative |
9 |
131 |
140 |
| Total |
99 |
131 |
230 |
Of the patients with positive blood cultures, 72 patients (63.2%) had Gram-positive bacterial sepsis, 41 patients (36.0%) had Gram-negative bacterial sepsis, and 1 patient (0.88%) had a mixed bacterial infection. The most common Gram-positive bacteria identified by blood culture were Staphylococcus coagulase negative and Staphylococcus aureus; the most common Gram-negative organisms identified were Klebsiella species and E. coli. There was no significant difference between the Gram stain result and the secondary outcomes of length of stay, lactate level, time to antibiotics, or mortality (P > 0.05).
DISCUSSION
In our patient cohort, the 92% correlation between having a positive blood culture and a positive PCR result was similar to prior literature reports. Previously, SeptiFastTM testing was determined to have a sensitivity of 87%, and a specificity of 85.8% for detecting sepsis pathogens.9 Concordance with blood culture was 86%9 despite the limit of detection at approximately 30 CFU/mL for most organisms.10 The PCR test result has potential implications for clinical predictions. Patients with positive sepsis PCRs on admission were more likely to have higher organ dysfunction scores (SOFA score) than those with negative PCRs.11 Rello et al. reported an almost linear relation between an increase in log numbers of pneumococcal copies (i.e., the genomic load) and the risk for septic shock and/or the need for mechanical ventilation.12
Our study provides some insight into our patient population and ICU diagnostic and therapeutic strategies. There are several limitations regarding the data in this study since only patients with a diagnosis of sepsis or septic shock based on ICD-10 codes with positive blood culture growth that triggered a subsequent PCR test were included in the sensitivity analysis. Specificity could not be calculated as the initial percentage of false positive results would have been zero since there were no cases in which individuals had a positive PCR with a negative blood culture. However, some bacteremic patients may have been missed through sample timing or the effect of prior antibiotics. In addition, PCR testing for bacterial identification using urine cultures, sputum cultures, or sites other than blood might help detect sepsis and characterize the infection. The study results did not include markers of disease severity, such as bilirubin and creatinine levels or platelet counts, to calculate the SOFA score and determine if the ICD-10 code diagnosis matched the clinical presentation.
Microorganism classification can help narrow the antibiotic spectrum, which potentially reduces the development of bacterial resistance, and can help predict the risk for complications and mortality.13 In our study, Gram-positive organisms were the most frequent cause of bacterial infection and accounted for 73.3% of infections in patients with severe sepsis and for 61% of patients with septic shock. In the United States, Gram-positive organisms are the most frequent cause of bacterial infection in septic patients.14 Overall, there is higher mortality with infections secondary to methicillin-resistant and methicillin-susceptible Staphylococcus aureus, Candida species, Pseudomonas species, and mixed pathogens.15 However, there is no difference in mortality between Gram-positive or Gram-negative negative bacteria in patients with septic shock.16 Li et al. have reported have reported that there was no difference in all-cause mortality, length of ICU stay, the requirement for mechanical ventilation, and requirement for renal replacement therapy in patients with culture positive sepsis or septic shock and in patients with culture negative sepsis or septic shock.17
Respiratory infections, specifically pneumonia, are the most common site of infection and are associated with highest mortality.18 In our patients, the most common infection sites in patients with severe sepsis included pulmonary (27%) followed by urinary and abdominal (8%). Similarly, in patients with positive PCR, pulmonary infections followed by urinary tract infections were the most common sites of infection, but there was higher frequency of multiple sites as the main category (76%).
The high mortality associated with severe sepsis and septic shock stresses the importance of early diagnosis and initiation of early goal directed therapy. Kumar et al. found that appropriate antibiotic therapy was started in 80% of septic shock cases studied, with overall survival rate at 43.7% (52% in those with appropriate antibiotics, 10.3% in those with inappropriate antibiotics).19 In this study on average, antibiotics were started in 4.4 hours in patients with no PCR testing and 3.0 hours in patients with positive PCR tests. The Surviving Sepsis Campaign most recent 2021 update recommends starting antibiotics ideally within the first hour of recognition of possible septic shock or sepsis.20 However, the mortality rate in this study was similar to published sepsis mortality outcomes. Overall mortality in the severe sepsis patients was 13% compared with reported 10%. Overall mortality in the septic shock patients was 37%, which matched prior literature reports of 40%.21
In summary, PCR testing had good diagnostic utility for the identification of bacterial infections, but positive test results were not associated with changes in the length of stay in patients from the ICU. Therefore, PCR testing should be used as a tool in conjunction with additional information, including bacterial classification by cultures and markers of disease severity. Prompt antibiotic initiation should contribute to better overall outcomes.
ACKNOWLEDGMENT
The Clinical Research Institute at Texas Tech University Health Sciences Center helped develop this project, including approval by the IRB.
REFERENCES
- Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence 2014;5(1):4–11. doi:10.4161/viru.27372
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315(8):801–810. doi:10.1001/jama.2016.0287
- Pletz MW, Wellinghausen N, Welte T. Will polymerase chain reaction (PCR)-based diagnostics improve outcome in septic patients? A clinical view. Intensive Care Med 2011;37(7):1069–1076. doi:10.1007/s00134-011-2245-x
- Stewart EJ. Growing unculturable bacteria. J Bacteriol 2012;194(16):4151–4160. doi:10.1128/JB.00345-12
- Lamoth F, Jaton K, Prod’hom G, et al. Multiplex blood PCR in combination with blood cultures for improvement of microbiological documentation of infection in febrile neutropenia. J Clin Microbiol 2010;48(10):3510–3516. doi:10.1128/JCM.00147-10
- Yanagihara K, Kitagawa Y, Tomonaga M, et al. Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit. Crit Care. 2010;14(4):R159. doi:10.1186/cc9234
- Huang, TD, Melnik E, Boaerts P, et al. Evaluation of the ePlex blood culture identification panels for detection of pathogens in bloodstream infections. J Clin Micro 2019;57(2):e01597–18.
- Timbrook TT, Morton JB, McConeghy KW, et al. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017;64(1):15–23. doi:10.1093/cid/ciw649
- Wellinghausen N, Kochem AJ, Disqué C, et al. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J Clin Microbiol 2009;47(9):2759–2765. doi:10.1128/JCM.00567-09
- Schaub N, Boldanova T, Noveanu M, et al. Incremental value of multiplex real-time PCR for the early diagnosis of sepsis in the emergency department. Swiss Med Wkly. 2014;144: w13911. Published 2014 Feb 4. doi:10.4414/smw.2014.13911
- Bloos F, Hinder F, Becker K, et al. A multicenter trial to compare blood culture with polymerase chain reaction in severe human sepsis. Intensive Care Med 2010;36(2):241–247. doi:10.1007/s00134-009-1705-z
- Rello J, Lisboa T, Lujan M, et al. Severity of pneumococcal pneumonia associated with genomic bacterial load. Chest 2009;136(3):832–840. doi:10.1378/chest.09-0258
- Cohen J, Cristofaro P, Carlet J, Opal S. New method of classifying infections in critically ill patients. Crit Care Med. 2004;32(7):1510–1526. doi: 10.1097/01.ccm.0000129973.13104.2d
- Martin GS, Mannino DM, Eaton S, et al. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 2003;348(16):1546–1554. doi:10.1056/NEJMoa022139
- Shorr AF, Tabak YP, Killian AD, et al. Healthcare-associated bloodstream infection: A distinct entity? Insights from a large U.S. database. Crit Care Med 2006;34(10):2588–2595. doi:10.1097/01.CCM.0000239121.09533.09
- Veterans Administration Systemic Sepsis Cooperative Study Group. Effect of high-dose glucocorticoid therapy on mortality in patients with clinical signs of systemic sepsis. N Engl J Med 1987;317(11):659–665. doi:10.1056/NEJM198709103171102
- Li Y, Guo J, Yang H, Li H, Shen Y, et al. Comparison of culture-negative and culture-positive sepsis or septic shock: a systematic review and meta-analysis. Crit Care 2021 May 8;25(1):167. doi: 10.1186/s13054-021-03592-8
- Esper AM, Moss M, Lewis CA, et al. The role of infection and comorbidity: Factors that influence disparities in sepsis. Crit Care Med 2006;34(10):2576–2582. doi: 10.1097/01.CCM.0000239114.50519.0E
- Kumar A, Ellis P, Arabi Y, et al. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest 2009;136(5):1237–1248. doi:10.1378/chest.09-0087
- Evans L, Rhodes A, Alhazzani W, et al. Executive summary: surviving sepsis campaign: international guidelines for the management of sepsis and septic shock 2021. Critical Care Med 2021 Nov 1;49(11):1974–82.
- Vincent JL, Jones G, David S, et al. Frequency and mortality of septic shock in Europe and North America: a systematic review and meta-analysis. Crit Care 2019;23(1):196. Published 2019 May 31. doi:10.1186/s13054-019-2478-6
Article citation: Solis X, Batson B, Morataya C, Iwuji K, Fairley H, Chakkour W, Nugent K, Islam E. The association of positive sepsis PCRs tests with patient mortality, length of stay, and antibiotic management in a medical intensive care unit. The Southwest Respiratory and Critical Care Chronicles 2023;11(49):1–9
From: Department of Internal Medicine, Texas Tech University Health Sciences Center, Lubbock, Texas
Submitted: 9/17/2023
Accepted: 10/1/2023
Conflicts of interest: none
This work is licensed under a Creative Commons
Attribution-ShareAlike 4.0 International License.